“Short read” sequencing through “clustering”
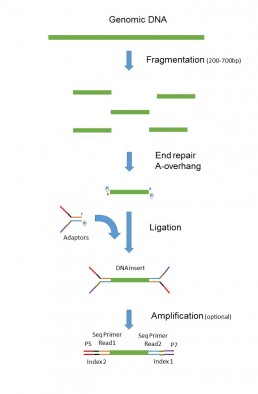
The library preparation for “paired-end” sequencing, according to the lllumina technology, consists in fragmenting the genomic DNA mechanically (Covaris, Bioruptor) or enzymatically (tagmentase) to sizes below 1 kb.
Indexed and complementary adapters of the sequencing primers are then ligated at each end of the DNA fragments to ensure sequencing from both ends (Figure 1).
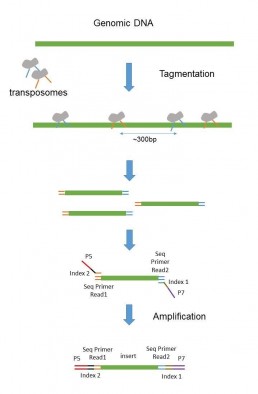
The technology using tagmentase, a modified transposase, fragments DNA and simultaneously adds sequencing adapters (Figure 2 : Nextera Technology).
A bead size selection step allows producing libraries of specific average insert size, which may be necessary for assembling particular genomic regions.
Sequencing by synthesis (SBS) begins with a clonal amplification step (clustering) and is carried out by a succession of cycles of incorporation of fluorescent dNTPs, signal detection, cleavage of the chromophore (see).