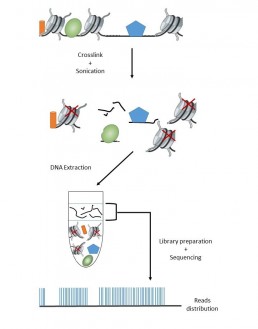
Mapping of chromatin accessibility sites: FAIRE seq
FAIRE seq (Formaldehyde-assisted isolation of regulatory elements) is a method for studying the accessibility of chromatin by nucleosome depletion. These regions of low nucleosome density, not bound to histones, are identified as genome regulatory areas (transcription factor binding site, promoters …).
The principle of the method is to use formaldehyde to covalently bind proteins and DNA, preferentially histones rather than genome-regulatory factors. After fragmentation by sonication, a Phenol chloroform extraction allows to recover the “free” DNA in the aqueous phase, proteins and nucleosomes being in the organic phase.
It is good practice to include a control sample as a reference of the whole genome, either by taking an aliquot after binding which would undergo a “decrosslink” at 65 ° C or from an aliquot which would go through all the steps of the protocol except the formaldehyde treatment (Rodrigez-Gil et al. 2018).
Rubriques associées
- Small RNA Sequencing
- Mapping of Transcription Start Sites – TSS
- TAPS/TAPSβ
- Enzymatic Methyl-seq (EM-seq™)
- Methylation of native DNA and RNA
- DNA binding sites map : CUT & RUN vs CUT & Tag
- Chromosome Contact map : 3C, 4C , 5C
- High Chromosome Contact map : HiC-seq
- Mapping of chromatin accessibility sites: DNase seq
- Indirect mapping of chromatin accessibility sites: MNase seq
- Mapping of chromatin accessibility sites: ATAC seq
- Mapping of RNA-protein interaction sites: CLIP seq
- Mapping of DNA-protein interaction sites: CHIP seq
- Mapping of DNA epigenetic marks: MeDIP
- Mapping of DNA epigenetic marks: Methyl seq
- BiSeq