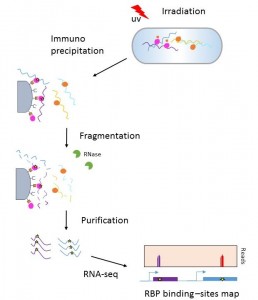
Mapping of RNA-protein interaction sites: CLIP seq
The analysis of RNA-protein interactions allows a better understanding of the post-transcriptional status of RNAs (their stability, localisation, circulation, translation level/efficiency, etc.). ). CLIP Seq (Crosss-Linking ImmunoPrecipitation Sequencing) is a method used to identify target sites for protein binding (PBR) on RNA. This technology combines the immunoprecipitation of relevant PBRs and high throughput RNA sequencing.
The principle of the methodology is based on covalently binding RBPs proteins to RNA by UV irradiation or enzymatic reaction. After cell lysis, RNA-protein complexes are isolated by immunoprecipitation with specific antibodies. The next step consists in RNA trimming. After ligation of the adaptors, protein removal and purification, RBPS binding sites are sequenced by RNAseq technology.
This complex protocol is detailed in C.Y. Flora et al. 2018
Rubriques associées
- Small RNA Sequencing
- Mapping of Transcription Start Sites – TSS
- TAPS/TAPSβ
- Enzymatic Methyl-seq (EM-seq™)
- Methylation of native DNA and RNA
- DNA binding sites map : CUT & RUN vs CUT & Tag
- High Chromosome Contact map : HiC-seq
- Indirect mapping of chromatin accessibility sites: MNase seq
- Mapping of chromatin accessibility sites: ATAC seq
- Mapping of DNA-protein interaction sites: CHIP seq
- Mapping of DNA epigenetic marks: MeDIP
- Mapping of DNA epigenetic marks: Methyl seq
- BiSeq