Methylation of native DNA and RNA
Several approaches to DNA methylation analysis have been developed to determine the hypo- or hypermethylated status of a large number of CpG islands, or even the entire genome. The most commonly used method is based on the bisulphite conversion of unmethylated cytosines to uracil, although it has a number of drawbacks, including the complexity of library preparation, conversion biases and the inability to distinguish between 5mC and 5hmC modifications.
Messenger RNA can also carry modifications (epitranscriptome), the most common of which found in eukaryotes is N6-Methyladenosine (m6A), that influences protein expression, by regulating splicing or sRNA stability, for example. The level of m6A is generally quantified by immunoprecipitation with some problems of specificity and yield (Deng X. et al. 2023).
For a more precise picture of the complexity of the genome and transcriptome, long fragment sequencing technologies (PacBio Hifi, Nanopore ONT) have enabled advances in the analysis of native DNA and RNA.
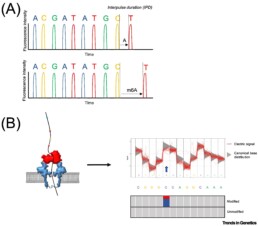
With these techniques, 5mC and 6mC modifications of DNA or RNA (without prior treatment altering the sample) are detected by a change in the sequencing signal (interpulse duration (IPD) or voltage difference). The state of methylation is measured by deviations from the canonical distribution which induces base-calling errors, localised, depending on the type of modification, at the level of the methylated nucleotides or adjacent positions. (see figure taken from van Dijk E.L. et al. 2023).
Analysis remains a tricky process. The software used depends on the detection approach chosen, whether it uses sequencing profiles or base calling errors. Since 2017, numerous developments have been undertaken in the bioinformatics tools as described by Furlan M. et al. 2021.
Rubriques associées
- Small RNA Sequencing
- Mapping of Transcription Start Sites – TSS
- TAPS/TAPSβ
- Enzymatic Methyl-seq (EM-seq™)
- DNA binding sites map : CUT & RUN vs CUT & Tag
- High Chromosome Contact map : HiC-seq
- Indirect mapping of chromatin accessibility sites: MNase seq
- Mapping of chromatin accessibility sites: ATAC seq
- Mapping of RNA-protein interaction sites: CLIP seq
- Mapping of DNA-protein interaction sites: CHIP seq
- Mapping of DNA epigenetic marks: MeDIP
- Mapping of DNA epigenetic marks: Methyl seq
- BiSeq