Ribo-Seq
RiboSeq is used to map and quantify the transcript footprints (~30nt) of functional ribosomes at a given time in a cell.
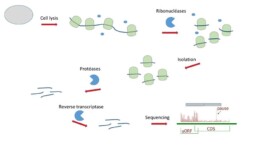
The protocol consists of first freezing the ribosomes on the mRNA with translation inhibitors. Then the sample is treated with ribonucleases to degrade any mRNA that is not protected by a ribosome. The mRNA-ribosome complexes are isolated. After treatment with proteases, the ‘protected’ mRNA fragments are purified, transcribed into cDNA and sequenced.
The identification of these regions allows the description of ribosome occupancy and mRNA translation efficiency (ratio between RiboSeq footprint count and RNAseq copy count of a given mRNA), alternative transcription sites, translation pause sites, differential genes expression at a post-transcriptional level, etc. (reviews: Ingolia N.T et al. 2018; Kage U et al. 2020)