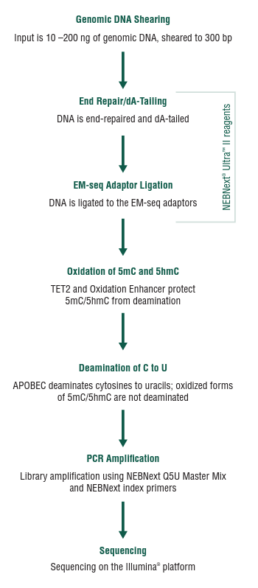
Enzymatic Methyl-seq (EM-seq™)
Bisulfite sequencing detects 5-methylcytosine (5mC) and 5-hydroxymethylcytosine (5hmC) at single-base resolution. However, bisulfite treatment damages DNA, which results in fragmentation, DNA loss, and biased sequencing data. To overcome these problems, enzymatic methyl-seq (EM-seq) was developed by NEB, see fig.1 (NEBNext® Enzymatic Methyl-seq Kit, NEB).
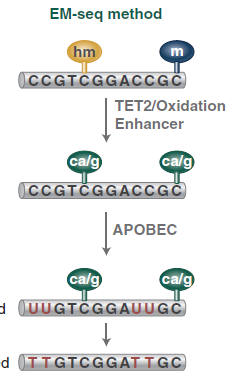
This method detects 5mC and 5hmC using two sets of enzymatic reactions (figure 2).
In the first reaction, TET2 (Ten-eleven-translocation 2 = methylcytosine dioxygenase) and T4-BGT (T4 Phage β-glucosyltransferase) convert 5mC and 5hmC into products that cannot be deaminated by APOBEC3A (Apolipoprotéins B mRNA editing enzyme, catalytic polypeptide-like). In the second reaction, APOBEC3A deaminates unmodified cytosines by converting them to uracils. Therefore, these three enzymes enable the identification of 5mC and 5hmC (Vaisvila et al., 2021).
Rubriques associées
- Small RNA Sequencing
- Mapping of Transcription Start Sites – TSS
- TAPS/TAPSβ
- Methylation of native DNA and RNA
- DNA binding sites map : CUT & RUN vs CUT & Tag
- High Chromosome Contact map : HiC-seq
- Indirect mapping of chromatin accessibility sites: MNase seq
- Mapping of chromatin accessibility sites: ATAC seq
- Mapping of RNA-protein interaction sites: CLIP seq
- Mapping of DNA-protein interaction sites: CHIP seq
- Mapping of DNA epigenetic marks: MeDIP
- Mapping of DNA epigenetic marks: Methyl seq
- BiSeq