Tn-Seq
Tn-Seq or sequencing of transposon-insertion libraries, which combines genome-wide transposon mutagenesis with high-throughput sequencing, helps assigning gene function in a wide range of bacterias and prokaryotes. By analysing the sites of transposon insertion, genes involved in survival functions (such as cell division or metabolism), stress response, virulence, etc. can be accessed (reviewed in Cain AK et al. Nat Rev. Genet. 2020).
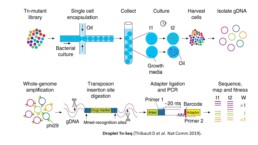
The basic principle of Tn-seq involves producing a library of mutant strains, each containing a unique transposon insertion. Two of the most commonly used transposons are Himar1 from the mariner family, which inserts randomly at TA dinucleotides, and Tn5, which can insert more broadly throughout the genome. When a transposon inserts itself in a gene, the gene’s function will be disrupted, affecting the fitness of the bacteria if the gene is essential or influential for its growth in specific conditions (ex: normal vs antibiotic exposure). After transposon-genome junction enrichment involving in MmeI digestion, subsequent adapter ligation and PCR amplification, the locations of the transposon insertions in the surviving strains are determined using high-throughput sequencing techniques. By comparing the distribution of insertion sites in the mutant strains to a reference genome, genes that are essential for survival under the conditions tested are identified.
Randomly barcoded transposon insertion sequencing (RB-TnSeq) modifies the initial TnSeq protocol by inserting unique 20-nucleotide barcode transposons enabling parallel mutant fitness profiling for several conditions in a single sequencing run (Wetmore KM et al. mBio 2015).
For eukaryote genome studies, this technic presents some limitation if using short read sequencing because of the numerous repeat regions of evolutionary or fitness importance. Therefore, mapping reads uniquely to repeated nucleotide sequences whose repeat units are longer than the sequence reads is compromised. Long read sequencing methods should be preferred (Yasir M et al. Sci. Rep 2022).
Droplet Tn-Seq (dTn-Seq) uses microfluidic methods of encapsulation to individualise transposon mutants enabling the study single cell growth, free from the influence of the other populations (Thibault D et al. Nat Comm 2019).
Platforms to contact for “mate pair” sequencing projects
Last update April 2025