Adaptive sampling
When doing large sequencing experiments, it can be hard to keep track of your region of interest in a rapidly expanding dataset. Isolating your strands of interest prior to sequencing can require time-consuming sample preparation.
Oxford Nanopore Technology (ONT) has developed a long-read sequencing method known as “Adaptive” sequencing.
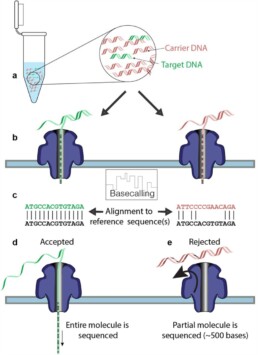
Thanks to adaptive sampling (AS), you can now skip time-consuming sample preparation and let the sequencer select your region of interest for you. During typical ONT sequencing, DNA strands are read via changes in electrical potential as the molecules transit through pores in a specially prepared membrane. Unlike Illumina sequencing, this ONT read data is available in real-time during the run, rather than only being available when the sequencing run is complete.
In AS, the sequence data is both generated and compared in real-time to that of a selected region of interest (ROI) within the genome. If the DNA strand entering the pore is not of interest, the real time data-streaming flags it up (the voltage is reversed) and the strand is ejected from the nanopore, leaving the pore free for the next strand in the sample.
As the strand of interest is processed, the system can quickly see that this is a target strand, and sequencing continues. This results in high coverage of sequencing data for your region of interest.
The user provides the FASTA sequence of the reference genome along with the coordinates of the ROI. Multiple non-overlapping different ROIs can be sampled simultaneously.
This technology allows for sample enrichment without the intensive and time-consuming library preparations required by, for example, Cas9 targeted enrichment for Nanopore sequencing. AS increases the proportion of low abundance reads in metagenomic samples (Martin et al. 2022) and has the potential for characterising regions of high variance (Weilguny et al. 2023).