Metatranscriptomics
Metatranscriptomics allows the analysis of messenger RNAs expressed within complex microbial communities, revealing functional activity rather than simply the genetic presence of microorganisms (Zang et al., 2021).
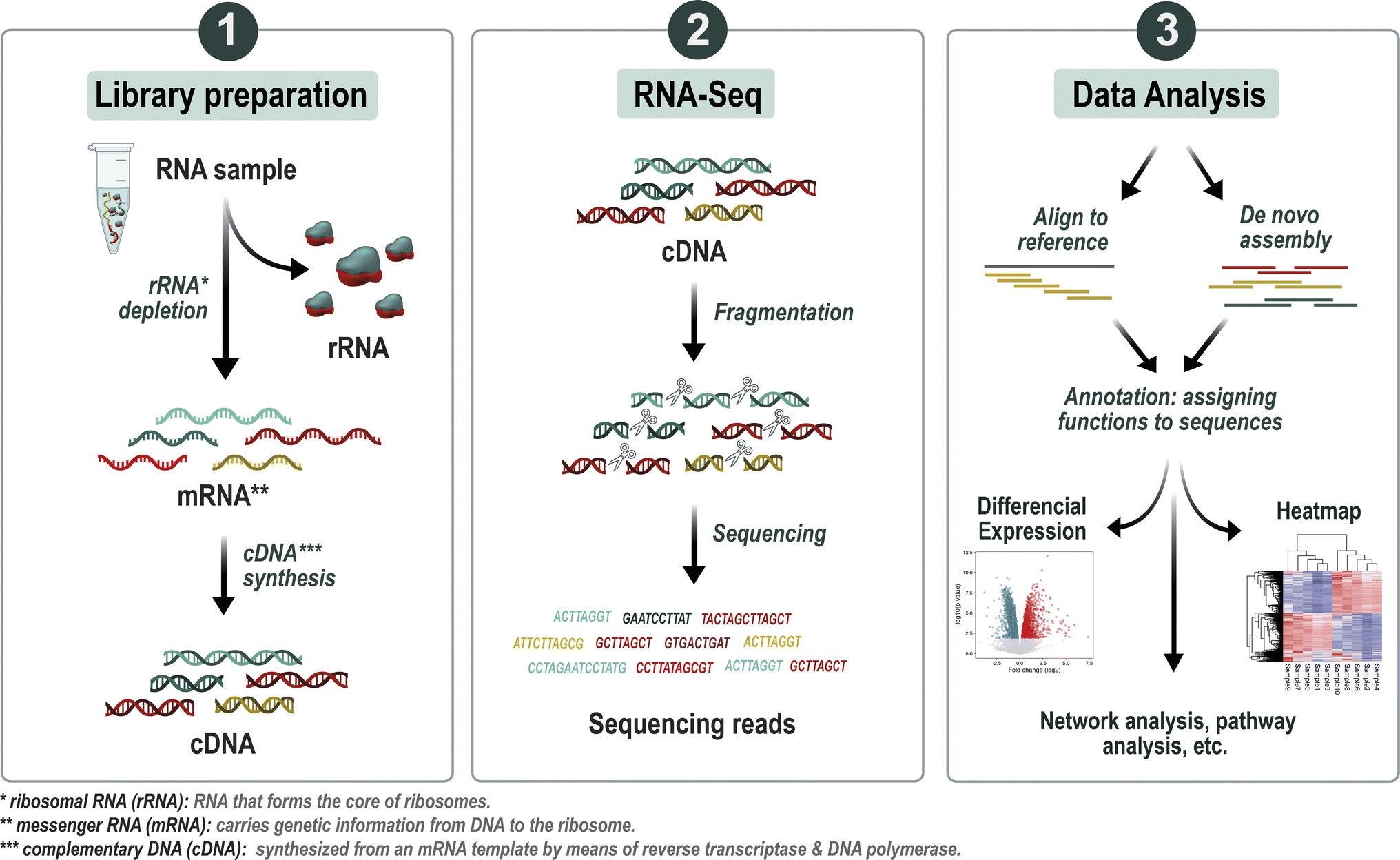
Experimental steps
a) Collection & extraction, sampling respecting rapid RNA stabilisation to prevent degradation. Extraction of total RNA from the sample (soil, water, intestinal flora, etc.), which mainly contains rRNA.
b) Depletion of rRNA (rRNA/tRNA) using commercial kits or bead capture, eliminating up to 70–85% of rRNA.
c) Fragmentation & library creation
- Conversion of mRNA to cDNA by reverse transcription with strand-specific protocols, often for Illumina platforms offering high-throughput short reads (≥100 million reads per sample), library preparation: adapter ligation, PCR amplification.
- Another option: direct RNA sequencing by nanopores (Oxford Nanopore), long reads, without PCR, without assembly bias, but with a higher error rate.
d) High-throughput sequencing, Illumina, Element BioSciences or MGI for short reads; Oxford Nanopore for long reads, providing access to complete transcripts or isoforms.
e) Bioinformatic analysis
- Quality control & filtering, using tools such as FastQC, Trimmomatic, SortMeRNA to remove low-quality reads or those derived from rRNA/tRNA.
- Assembly/mapping, assembly of reads (de novo or by mapping to reference databases), taxonomic assignment using tools such as Kraken2, BLASTN, mapping to SILVA, Greengenes or genomic databases.
- Functional annotation, identification of ORFs, annotation via KEGG, MetaHIT, M5nr, COG, GO, VFDB or CARD databases depending on the request (antibiotic resistance, virulence, etc.) → assignment of biological functions.
New technique: Use of direct RNA sequencing by nanopore to access long transcripts, isoforms and structural RNAs without cDNA, currently being optimised.
Zhang Y, et al. Metatranscriptomics for the Human Microbiome and Microbial Community Functional Profiling, Annual Review of Biomedical Data Science Vol. 4:279-311 (juillet 2021) https://doi.org/10.1146/annurev-biodatasci-031121-103035
Esser M, Brinkmann M and Hecker M, Solving freshwater conservation challenges through next-generation sequencing approaches, DOI: 10.1039/D4VA00112E (Perspective) Environ. Sci.: Adv., 2024, 3, 1181-1196, https://pubs.rsc.org/en/content/articlehtml/2024/va/d4va00112e