UCA GenomiX traque les variants SARS-CoV-2 dans les eaux usées
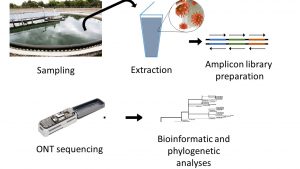
La plateforme UCA GenomiX (IPMC), a développé depuis plusieurs mois une méthode pour mesurer les niveaux des différentes formes du virus du SARS-CoV-2 qui circulent dans les eaux usées de la métropole niçoise. La détection de taux élevé de SARS-CoV2 dans les selles de patients a en effet suggéré l’intérêt d’y suivre ses niveaux dans les stations d’épuration. L’apparition de nouveaux variants rend particulièrement intéressante l’approche par séquençage, qui permet de les détecter facilement. La lecture de l’intégralité du génome viral (30000 nucléotides) est effectuée par une technique de séquençage Oxford Nanopore, sur une série d’amplicons produits à partir d’extraits d’eaux usées. En septembre 2020, UCA GenomiX a commencé a développé un partenariat avec la Ville de Nice pour réaliser le séquençage d’échantillons collectés dans une vingtaine de quartiers différents, réalisant ainsi une quantification précises des différents variants présents sur le territoire. Cette collecte par quartier a permis d’alerter très tôt la Métropôle niçoise sur la circulation de certains variants “à risque” dans certains quartiers de la ville. Tout en poursuivant cette surveillance, l’équipe collabore désormais avec la Société Véolia qui souhaite industrialiser cette approche, en s’appuyant sur l’expertise développée par UCA GenomiX. La réalisation des premières analyses a en effet réclamé la construction d’une chaîne complète de traitement, allant de la récupération des échantillons jusqu’à la production des banques, leur séquençage, et leur analyse bioinformatique. Cette initiative a été suivie par d’autres, comme celle de l’observatoire Ôbépine, (https://www.reseau-obepine.fr/), afin de l’élargir à l’ensemble du territoire français
Les personnes de l’IPMC impliquées dans le projet : Géraldine Rios, Assistant Ingénieur CNRS ; Rainer Waldmann, chercheur INSERM ; Caroline Lacoux (équipe Bernard Mari), chercheuse CNRS ; Kévin Lebrigand, ingénieur CNRS ; Anna Diamant, étudiante Master; Pascal Barbry, DR CNRS.

