Single-cell atlas of the whole human lung
First integrated Human Lung Cell Atlas reveals insights into lung diseases
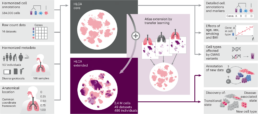
The largest and most comprehensive cell map of the Human Lung was recently published in Nature Medicine (Link). Revealing the great diversity of cell types in the lung and key differences between health and disease, the Human Lung Cell Atlas will be a valuable resource for lung researchers.
By combining data from nearly 40 studies, researchers created the first integrated single-cell atlas of the lung, revealing rare cell types and highlighting cellular differences between healthy people. In addition, the study found common cell states between lung fibrosis, cancer and COVID-19, offering new ways of understanding lung disease, which could help identify new therapeutic targets.
The Human Cell Atlas Lung Biological Network is a group of scientists who collaborate to map the cell types and states present in human airways. This group is coordinated among others by Pascal Barbry, head of UCAGenomiX, Nice and France Génomique has contributed to the project.