Plateforme de séquençage de Paris-Saclay
The Paris Saclay Transcriptomics Platform (POPS), located in Gif sur Yvette (91), was created in 2003 with the primary mission of offering a range of technical and bioinformatics services for the analysis of plant transcriptomes. It is a ‘customised integrated service’.
POPS enables the public and private scientific community to answer biological questions through high-throughput RNA sequencing, commonly known as RNA-Seq. We carry out research projects in collaboration with national and international research teams. We have chosen to specialise exclusively in plants in order to develop strong expertise and provide our partners with the best possible advice on their projects. To date, we have worked on more than 93 different species.
Our field of application requires us to remain at the cutting edge of technology in the field of sequencing and data analysis in order to offer technical protocols and bioinformatic analyses that are both biologically and financially relevant and interesting. Our second mission is ‘development’.
Finally, it is important to pass on the knowledge acquired through the community. The platform provides training, participates in teaching, co-organises ‘plant omics days’ and welcomes interns. This is an essential aspect called ‘Training & Scientific Animation’.
Services
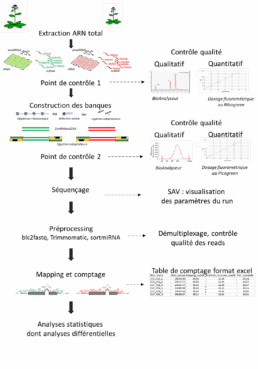
With its wealth of expertise in plants, POPS offers various protocols and analyses to answer users’ biological questions. The different stages involved in carrying out a project are as follows:
(*These stages can be handled by our collaborators.)
- Discussion of the experimental design and selection of the most appropriate technology,
- Grinding of samples*,
- RNA extraction*,
- Qualitative and quantitative control of RNA,
- Construction of libraries,
- Qualitative and quantitative control of libraries,
- Sequencing,
- Data analysis*,
- Interpretation of results*,
- Submission to databases*.
Expertise
Please feel free to contact them for customisable protocols tailored to your specific needs.
- Sequencing of polyadenylated and non-polyadenylated RNAs
- Sequencing of small RNAs: miRNA, siRNA, etc.
- Sequencing of long fragments (collaborative projects)
- Microtranscriptomics or sequencing of very small amounts of RNA (from 75 pg of total RNA).
- Metatranscriptomics or sequencing of the transcriptome of several species from the same sample.
- Fingerprinting
- Single cell (10X, Rhapsody, ScaleBiosciences)
- Large-scale transcriptomic screening (BRB-Seq)
Bioinformatics and statistics
POPS services can include bioinformatics and statistical analyses of the data generated. This is made possible thanks to the involvement of the IPS2 ‘Genomic Networks’ team, known for its development of methods and pipelines for omics data.
Bioinformatics and statistical analysis performed by POPS ensures that data remains private for a period agreed upon with the collaborator and is then made public through deposit in the CATdb database (website) and the international NCBI database (GEO/SRA).
As part of its ‘integrated service’, POPS systematically offers pre-processing to ensure data quality and storage for 6 months after sending to collaborators (up to a limit of 1 TB). This service can be supplemented by other services such as:
- Bioinformatic analysis per sample to provide a count table
- Bioinformatic and statistical analysis of conventional RNA-seq (differential analyses, co-expression analyses, enrichment tests, study of cis-regulatory elements of transcription)
- Bioinformatic and statistical analysis of single-cell data (identification of cell clusters, biomarkers and differential analyses)
- Bioinformatic and statistical analysis for transcriptomic screening.
- Construction of a reference transcriptome (reference genome not accessible) and bioinformatic and statistical analysis
- De novo transcriptome assembly and annotation (including Nanopore)
Equipments
- Quality control: Bioanalyzer (Agilent), Fragment Analyser (Agilent), CLARIOstar Plus (BMG Labtech), Qubit (Invitrogen), Pippin HT (Sage Science)
- Cell sorter: MACSQuant tyto (Miltenyo Biotec)
- Cell separator: Chromium (10X Genomics), HT Express Rhapsody (BD Sciences)
- Pipetting robot: Biomek FX (Beckman)
- Sequencers: NextSeq500 (Illumina), NovaSeq (external service providers), MinION (Nanopore), P2 (Nanopore)
- Sonicator: M220 Covaris (LGS Genomics)
- IT: Server (DELL), Computing station (DELL)
Main achievements
Last update: Aug 2025

Certification / Quality assurance
ISO9001 certified since 2012
Labels
IBiSA platform since 2008
National Strategic Platform by INRAE since 2008



Platform Managers
Scientific Manager
Etienne Delannoy
Operations Manager
Médine Benchouaïa
IPS2, Institute of Plant Sciences – Paris-Saclay
Building 630, avenue des Sciences
Plateau du Moulon
91190 – Gif-sur-Yvette
Contacts
Etienne Delannoy,
Médine Benchouaïa
pops.ips2@universite-paris-saclay.fr
POPS, Plateforme TranscriptOmique de l’iPS2
01 69 15 77 19

